No association between BCR-ABL1 fusion genes and clinical features of acute lymphoblastic leukaemia in Ghanaian patients
Victor Obeng Ofori, Amma Benneh-Akwasi Kuma, Susan Crocker, Edeghonghon Olayemi
Corresponding author: Edeghonghon Olayemi, Department of Haematology, University of Ghana Medical School, Accra, Ghana 
Received: 13 Apr 2020 - Accepted: 30 May 2020 - Published: 08 Oct 2020
Domain: Internal medicine,Oncology
Keywords: BCR-ABL1 fusion gene, acute lymphoblastic leukaemia, clinical and laboratory features
©Victor Obeng Ofori et al. PAMJ Clinical Medicine (ISSN: 2707-2797). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Victor Obeng Ofori et al. No association between BCR-ABL1 fusion genes and clinical features of acute lymphoblastic leukaemia in Ghanaian patients. PAMJ Clinical Medicine. 2020;4:57. [doi: 10.11604/pamj-cm.2020.4.57.22829]
Available online at: https://www.clinical-medicine.panafrican-med-journal.com//content/article/4/57/full
Research 
No association between BCR-ABL1 fusion genes and clinical features of acute lymphoblastic leukaemia in Ghanaian patients
No association between BCR-ABL1 fusion genes and clinical features of acute lymphoblastic leukaemia in Ghanaian patients
Victor Obeng Ofori1,&, Amma Benneh-Akwasi Kuma2, Susan Crocker3, Edeghonghon Olayemi2,&
&Corresponding author
Introduction: The BCR-ABL1 fusion gene has been associated with poor prognosis in acute lymphoblastic leukaemia (ALL). This study was designed to determine the presence, frequency and associated laboratory and clinical features of the BCR-ABL1 gene in Ghanaian patients diagnosed with ALL.
Methods: this was a cross-sectional study using archival methanol-fixed bone marrow aspirate slides of morphologically diagnosed ALL patients. Presence of the BCR-ABL1fusion gene was determined by fluorescent in situ hybridization. Clinical features and haematological parameters were extracted from the patients´ medical records.
Results: seventeen patients were studied, 13 (76.5%) males and 4 (23.5%) females. Median age was 24 years (range 15 to 67 years). A frequency of 29.4% was obtained for the BCR-ABL1 fusion gene. There was no significant association between presence of BCR-ABL1 and selected clinical features (lymphadenopathy, splenomegaly and hepatomegaly). All patients had moderate to severe anaemia with median haemoglobin concentration of 7.6 g/dL (range 3.7 to 8.7 g/dL). Median haemoglobin concentration for BCR-ABL1 positive patients was higher than that for negative patients (7.6 vs. 7.4 g/dL, p = 0.506); who also had higher median white blood cell counts (24.76 vs. 13.02 X 109/L), but lower median platelet counts (58.0 vs. 64.5 X 109/L, p = 0.721) and bone marrow blast percentages (78.5 vs. 98.0%, p = 0.851) compared to negative patients.
Conclusion: BCR-ABL1 fusion gene was detected in nearly one third of adult ALL patients in this study, with no significant association with common haematological parameters and clinical features of the disease.
Acute lymphoblastic leukaemia (ALL) is characterised by the presence of more than 20% leukaemic blasts in the bone marrow and specific leukaemic cytogenetic or/and molecular genetic abnormalities [1]. The chimeric BCR-ABL1 fusion gene results from translocation of the ABL cellular oncogene on chromosome 9 to the BCR gene on chromosome 22 [2]). This results in synthesis of either a 210 kD or 190 kD protein with enhanced tyrosine kinase activity compared with the normal 145 kD protein [3,4]. The frequency of the BCR-ABL1 fusion gene in patients with ALL ranges from 1-5% to 11-29% in children and adults respectively [5]. The two main types of the BCR-ABL1 fusion transcripts (p190 and p210) occur in ALL but the p190 is more prevalent [6,7]. In a study of 56 adult ALL patients in the United States by the Cancer and Leukaemia Group B (CALGB), the p190 variant accounted for 77% of patients [8]. The BCR-ABL1 fusion gene is a poor prognostic indicator in ALL and it has been associated with decreased overall and event free survival rates [9,10], with varying frequencies across different populations. The presence and frequency of this mutant gene in Ghanaian ALL patients is unknown, patients diagnosed with ALL in Ghana are not routinely screened for its presence. Treatment is therefore not modified to include tyrosine kinase inhibitors (TKIs), which are available for those who may have the mutation. Their incorporation in to therapeutic protocols in other parts of the world has been associated with improved survival in BCR-ABL1 positive ALL [11,12]. This study was designed to determine the presence, frequency and the associated laboratory and clinical features of the chimeric BCR-ABL1 gene in patients diagnosed with ALL at the Department of Haematology, Korle Bu Teaching Hospital (KBTH).
Study design: this was a cross-sectional study using methanol-fixed archived bone marrow slides of patients diagnosed with ALL.
Study site: the study was carried out at the Department of Haematology, Korle Bu Teaching Hospital. The department provides laboratory and clinical services to patients with various haematological disorders from all over Ghana and neighbouring West African countries. About 4,800 patients are seen in the department each year, with about 10 patients diagnosed with ALL per year. Further laboratory analysis with fluorescent-in situ hybridisation (FISH) was carried out at the Queen´s Laboratory for Molecular Pathology at Queen´s University, Canada.
Study population: the population consisted of ALL patients diagnosed at the study site from January 2013 to May 2017. Patients who were 15 years or older, with morphologically diagnosed ALL for whom unstained, fixed, bone marrow aspirate slides were available were included in the study. Twenty five of the 37 ALL patients seen within the study period met the inclusion criteria and were selected for the study.
Selection of samples and data collection: unstained bone marrow aspirate slides of patients that qualified for the study were retrieved from storage using laboratory numbers obtained from the haematology laboratory log book. A data abstraction form was used to obtain the following information from the patients´ medical records:clinical features at presentation: hepatomegaly, splenomegaly, lymphadenopathy and/or presence of mediastinal mass; the following laboratory variables: haemoglobin (Hb) concentration, white cell and platelet counts were obtained from full blood count (FBC) performed at the time bone marrow aspirate was taken for diagnosis. The blast cell percentages were obtained from morphology report of bone marrow aspirate smears.
Materials and methods: fluorescence in-situ hybridisation technique was used for the detection of the BCR-ABL1 fusion gene in the archived bone marrow aspirate slides.
Procedure: the fluorescent in-situ hybridisation technique was performed using the protocol at Queens Laboratory for Molecular Pathology (Queens University, Canada) as described below.
Preparation of positive and negative control smears: a positive control smear was prepared from a cell culture of a commercially prepared BCR-ABL1positive cell line (K-562 (ATCC ® CCL -243)). A peripheral blood smear prepared from an anonymous BCR-ABL1 negative volunteer provided by the Queens Laboratory for Molecular Pathology was used as a negative control slide. Both slides were fixed in methanol for 3 minutes.
DNA unmasking: the slides were immersed in methanol for 1 minute followed by incubation in 2 x Saline-Sodium Citrate (SSC) at room temperature for 5 minutes. The slides were then incubated in 0.2N HCl for 5 minutes. A total of 500 uL of pepsin was added to the pre-warmed 49.5 mL of 0.01N HCl in water bath at 37°C. The resulting mixture was mixed well. The slides were taken from the 0.2N HCl solution and excess liquid removed with a paper towel and immediately immersed into the pepsin/HCl solution incubating at 37°C for 7 to 15 minutes. The slides were washed in double distilled H2O for 10 minutes followed by fixation in 1% formaldehyde for 5 minutes, immersion in 1% phosphate buffered saline for 5 minutes and sequential dehydration in 70%, 85% and 100% ethanol for 2 minutes in each solution. The slides were observed under phase contrast microscope for digestion progress (unfinished digestion is signified by the appearance of white shiny cells with undefined nuclei whereas cells with clear blue nuclei shows complete digestion). If digestion was not finished, the slides were re-immersed in pepsin/HCl solution for a longer period and the subsequent steps followed until the phase contrast microscope shows finished digestion.
Denaturation and hybridization: the slides were air dried completely prior to the addition of the probe in the hybridisation steps which is described below: the ThermoBrite Denaturation/Hybridisation system was turned on and the program set for the following parameters: denaturation time: 2 minutes; denaturation temperature: 73°C; hybridisation time: 24 hours; hybridisation temperature: 37°C. The DNA probe (Vysis LSI BCR/ABL DC/DF translocation probe), Vysis LSI/WCP hybridisation buffer and the purified water were removed from storage and the reagents allowed to reach room temperature. The BCR/ABL DNA probe and the hybridisation buffer were vortexed for 2 to 3 seconds followed by centrifugation for 2 to 3 seconds. Seven microlitres (7 uL) of the hybridisation buffer, 2 uL of purified water and 1uL of the BCR/ABL DNA probe were transferred into a micro-centrifuge. The mixture was vortexed and centrifuged for 2 to 3 seconds each. A micropipette was used to apply 10 uL of the probe mixture to the target area of each slide. A cover slip was immediately applied without introducing bubbles. The coverslip was sealed using a syringe filled with rubber cement. Two ThermoBrite humidity cards saturated with distilled water were inserted into the slot positions in the unit lid of the ThermoBrite hybridisation/denaturation system. The slides were placed on the heating surface of the ThermoBrite Hybridisation/Denaturation system when prompted and was ensured that, the frosted edge of the slide hanged over the heating surface, lay flat and properly aligned into the marked positions in the slide locator. The ThermoBrite lid was closed and the program was started for denaturation and hybridisation to occur overnight.
Post-hybridisation wash: the post-hybridisation wash was carried out according to the following procedure: The room was darkened and the coverslip removed from the slide by peeling off rubber cement. The slides were incubated in 2x SSC/0.3% IgePal solution (180 uL IgePal in 60 mL 2 x SSC) at 73°C for 2 minutes. The temperature was increased by 0.5°C for each slide if the slides were more than one. The slides were washed in 2x SSC for 5 minutes and air-dried in upright position under foil cap.
Counterstaining: counterstaining was carried out by the application of 10 uL DAPI to the middle of each slide. Coverslip was applied and air bubbles pushed out. They were stored in the dark at - 20°C until fluorescent microscopy was carried out in the following step.
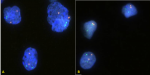
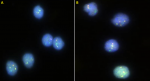
Fluorescent microscopy: the fluorescent microscopy room was darkened and immersion oil added to the slides. They were then observed under the fluorescent microscope using the spectrum orange, spectrum green and the dual filter which allows the visualisation of ABL1, BCR and BCR/ABL1 gene respectively. A total of 100 interphase nuclei were scored for each slide. Images of the slides were captured using the imaging software GenASIs FISH view and processed using Case Data Manager (CDM). The cut-off point for BCR-ABL1positivity was 3% and 15% for double fusion and single fusion respectively (values for fluorescent signals were compare to normative cut-off database at the QLMP). Representative images are shown in Figure 1 and Figure 2.
Data handling: study cases were assigned unique identification numbers, these were used to label the respective bone marrow aspirate slides and also for subsequent data processing. Data was stored on a password protected computer. The names of subjects were not used in the data but were however kept in a different file. Only the Investigators had access to data obtained from the study.
Statistical analysis: the data was entered into Microsoft Excel and exported to Statistical Package for the Social Sciences (SPSS version 20) for analysis. Data was expressed using summary and descriptive statistics such as frequency, percentages and median as appropriate and presented in tabular form. Chi Square, Fisher exact test and non-parametric test (Mann-Whitney U-test) were used to assess the association between categorical factors. The study received approval from the ethical and protocol review committee, College of Health Sciences, University of Ghana, No. CHS-Et/M.S-P 4.2/2016/2017.
Bone marrow aspirate slides from 25 patients morphologically diagnosed as ALL were available for study. FISH was successfully carried out on 17/25 (68%) slides as 8/25 (32%) slides were not suitable for processing due to poor storage and deterioration.
FISH results and frequency of the BCR-ABL1 gene in samples: five out of seventeen patients (29.4%) were positive for the BCR-ABL1 gene. Of the 5 positive cases, 4 were double fusion and one had single fusion. The percentage scores for positive cases of the gene were 46%, 37%, 9% and 8% for double fusion and 20% for single fusion (Table 1).
Selected FISH images: Figure 1 shows a fluorescent photomicrograph of the negative control slide (A) and a negative patient (B). No fusion signals are present. Figure 2 shows the fluorescent photomicrograph of a positive control slide (A) and a positive patient slide (B). Double fusion signals which appear yellow are seen in the lower left and topmost nuclei (A). Double fusion signals (either yellow dots or orange and green dots in juxtaposition) are seen in the two nuclei in the middle (B).
Key to interpretation of images: BCR-GENE (locus 22q11.2) - green signals (dots); ABL GENE (locus q34.1) - orange signals (dots); BCR-ABL1 fusion gene - orange and green signals in juxtaposition or yellow signal are seen for BCR-ABL1 positive cases. Double fusion cases have 2 signals whereas 1 signal is seen single fusion cases or may represent proximity of signals due to chance). BCR-ABL negative cases show separated green and orange signals but no fusion signals).
Demographics
In the study 76.5% (13/17) of the samples were from males and 23.5% (4/17) were from females. Positive cases consisted of ¼ (25%) of females and 4/13 (30.8%) of males. The ages of participants ranged from 15 years to 67 years (median age 24 year). Participants in the study were categorized into two groups; adolescent and young adult group (AYA group, 15 to 39 years) and older adult group (40 years and older) respectively based on the guidelines of the U.S. National Comprehensive Cancer Network (NCCN) [13]. All the positive participants were in the AYA group 5/12 (41.7%).
Descriptive and Inferential statistics of clinical features and BCR-ABL1 Gene: Table 2 show the frequencies of clinical features and test of association with the BCR-ABL1 gene.
Descriptive and inferential statistics of haematological parameters: association between BCR-ABL1 Fusion Gene and Haematological Parameters There was no significant difference (using the Mann-Whitney-U test) in the median of the following haematological parameters: WBC count (13.02 vs. 24.76 X 109/L), P = 0.879), Hb concentration 7.4 vs. 7.6 g/dl, P = 0.506), blast percentage (94% vs. 78.5%, p= 0.851 and platelet count (64.5 vs. 58 X 109/L p = 0.721 ) between patients negative or positive for the BCR-ABL1 fusion gene.
The BCR-ABL1 fusion gene was present in nearly one-third of adolescent and adult patients in our study. Additionally, our findings suggested no statistically significant association between the presence of the gene and clinical features or haematological laboratory parameters. The gene frequency of 29.4% for BCR-ABL1 in this study is consistent with the rates of 28.3% reported from India by Chopra et al. [14], and 11-29% reported by Mrozek et al. [15] in a review of adult ALL studies from USA, United Kingdom and France. However, it is higher than a prevalence of 12.5% obtained by Ajuba et al. [16] in Nigeria. The difference between this study and that of Ajuba et al. (both conducted in West Africa) may arise from the fact that our study excluded children since the BCR-ABL1gene is more common in adults Mrozek et al. [15]. Racial differences may account for some of the disparities seen. For example, in Saudi Arabia, none of the 16 adult ALL patients investigated by El-Sissy et al. were positive for the gene [17] while Ariffin et al. in Singapore demonstrated variation in frequency between Indians, Malays and Chinese [18].
Age and gender: All the BCR-ABL1positive patients were in the (AYA) group, the exact reasons for this are not clear; although globally, ALL is commoner in children and adolescents. Ghana has a young population with almost 60% less than 24 years old and only 11% older than 50 years. This contrasts with the population structure of advanced countries such as the United States, Germany and Japan with relatively older populations where about 33%, 25% and 23% respectively are less than 24 years old whereas 34%, 40% and 45% are older than 50 years [19]. Thus, the lower proportions of the elderly in this study may reflect the relatively shorter life expectancy at birth (61.3 years in Ghana) compared to that of the United States, Germany and Japan which is 78.9, 80.8 and 83.6 years respectively [20].
Clinical feature: lymphadenopathy was present in 41.2% of all the patients in this study of which 14.3% were BCR-ABL1 positive whereas 23.5% of the patients studied had splenomegaly of which a 25% was BCR-ABL1 positive. Hepatomegaly was evident in 35.7% of study participants with 20% showing BCR-ABL1 positivity, no patient had a mediastinal mass. The frequencies of organomegaly are lower than reported in a study by Elbossaty et al. in Egypt in which lymphadenopathy, splenomegaly and hepatomegaly occurred in 58, 58 and 54% of adult all patients [21]. However, it is consistent with the findings in Netherlands by Daenen et al. in which organomegaly (lymphadenopathy, splenomegaly and hepatomegaly) were less prevalent with a combined frequency of 40% in adult ALL [22]. As reported in our study, Westbrook et al. in a study by the Cancer and Leukaemia Group B in USA found no statistically significant association between BCR-ABL1 positivity and these clinical features [8].
Haematological parameters: the mean white blood cell (WBC) count, bone marrow blast percentages and platelet counts were lower in BCR-ABL1 positive participants than those who were negative. Contrastingly, the mean Hb concentration for BCR-ABL1 positive participants was higher than in participants negative for the gene. These were in contrast to the findings by Gleißner et al. in Germany in which BCR-ABL1 positive patients had statistically significantly higher WBC count and Hb concentration than BCR-ABL1 negative patients [6]. However, similar to Gleißner et al. our study found no statistically significant difference in white cell count, platelet count, Hb concentration and blast percentage in BCR-ABL1positive and negative patients [6]. All the patients in this study had severe to moderate anaemia with Hb concentration ranging from 3.7 to 8.7 g/dL. Also, with the exception of one, all patients in the study (approximately 94%) had thrombocytopenia. These frequencies and degree of anaemia and thrombocytopenia is higher compared to earlier reports from Denmark and Italy [23,24]. This may result from delayed presentation of our patients. This challenge is common in developing countries such as Ghana due to inadequate number of health facilities as well as haematologists. Secondly, the prevalence of anaemia is higher in Ghana compared to high income countries. For instance, the proportion of non-pregnant women with Hb concentration below 12 g/dL in Ghana is 56% whereas in Canada, Germany, Japan and the United States of America it is 16, 18, 22 and 12% respectively [25]. The clinical outcome of most of the participants in our study could not be determined. Due to the fact that most of the patients studied 13/17 (76.5%) were lost to follow up. Our study was limited by the number of properly stored bone marrow slides available for study. About a third of the marrow slides were not suitable for processing due to poor storage.
We describe for the first time the detection of the BCR-ABL1 fusion gene in Ghanaian ALL patients. With a frequency of almost 30%, the BCR-ABL1 fusion gene is an important molecular genetic lesion in adolescent and adult ALL cases in our environment. However, it has no significant association with the clinical features and haematological parameters of the disease. We recommend a larger multicentre prospective study of BCR-ABL1gene in both children and adults with ALL involving the characterization of the associated molecular signatures such as the IKZF1 and the PAX genes.
What is known about this topic
- The BCR-ABL fusion gene is a poor prognostic indicator in ALL;
- Incorporation of targeted therapy with TKIs improves prognosis;
- The frequency of the BCR-ABL 1 fusion gene frequency varies across different populations.
What this study adds
- The detection of the BCR-ABL1fusion gene is described for the first time in Ghanaian ALL patients;
- The gene was only seen in adolescents and young adults;
- From our findings there is no statistically significant association between presence of the gene and clinical features or haematological laboratory parameters.
The authors declare no competing interests.
VOO, ABAK and EO designed the study. VOO carried out the bench work under the supervision of SC. VOO and EO analysed the data. VOO and EO wrote the first draft. All authors participated in writing subsequent drafts, revised it critically and approved the final version before submission.
We acknowledge the assistance of Brooke Ring-Snetsinger, Shakeel Virk and Andy Zhang of Queens Laboratory for Molecular Pathology for their assistance. We are also grateful to Francisco Torto who retrieved the patients´ clinical files as well as Dr. Tom Ndanu (School of Dentistry, University of Ghana) for his assistance with statistical analysis.
Table 1: features of BCR-ABL1 positive patients
Table 2:
association between BCR-ABL1 fusion gene and clinical features
Figure 1: fluorescent
photomicrograph of a negative control slide (A) and a negative patient slide
(B)
Figure 2: fluorescent photomicrograph of a positive control slide (A) and a positive patient slide (B)
- Hoffbrand V, Moss PAH. Hoffbrand's Essential Haematology. 7th edition Hoboken Wiley. 2015.
- Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3407-15. PubMed | Google Scholar
- Kumar V, Abbas AK, Fausto N, Aster JC. Robbins and Cotran Pathologic Basis of Disease. 8th edition Philadelphia.Elsevier. 2009.
- Hoffman R, Benz EJ, Silberstein LE, Heslop H, Weitz J, Anastasi J. Hematology: basic Principles and Practice. 6th edition Philadelphia. Elsevier. 2012.
- Mrozek K, Harper DP, Aplan PD. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Hematology/oncology clinics of North America. 2009;23(5):991-1010. PubMed | Google Scholar
- Gleißner B, Gökbuget N, Bartram CR, Janssen B, Rieder H, Janssen JW et al. Leading prognostic relevance of the BCR-ABL translocation in adult acute B-lineage lymphoblastic leukemia: a prospective study of the German Multicenter Trial Group and confirmed polymerase chain reaction analysis. Blood. 2002;99(5):1536-43. PubMed | Google Scholar
- Cimino G, Pane F, Elia L, Finolezzi E, Fazi P, Annino L et al. The role of BCR/ABL isoforms in the presentation and outcome of patients with Philadelphia-positive acute lymphoblastic leukemia: a seven-year update of the GIMEMA 0496 trial. Haematologica. 2006;91(3):377-80. PubMed | Google Scholar
- Westbrook CA, Hooberman AL, Spino C, Dodge RK, Larson RA, Davey F et al. Clinical significance of the BCR-ABL fusion gene in adult acute lymphoblastic leukemia: a Cancer and Leukemia Group B Study (8762). Blood. 1992;80(12):2983-90. PubMed | Google Scholar
- Pullarkat V, Slovak ML, Kopecky KJ, Forman SJ, Appelbaum FR. Impact of cytogenetics on the outcome of adult acute lymphoblastic leukemia: results of Southwest Oncology Group 9400 study. Blood. 2008;111(5):2563-72. PubMed | Google Scholar
- Fletcher JA, Lynch EA, Kimball VM, Donnelly M, Tantravahi R, Sallan SE. Translocation (9; 22) is associated with extremely poor prognosis in intensively treated children with acute lymphoblastic leukemia. Blood. 1991;77(3):435-9. Google Scholar
- Brissot E, Labopin M, Beckers MM, Socié G, Rambaldi A, Volin L et al. Tyrosine kinase inhibitors improve long-term outcome of allogeneic hematopoietic stem cell transplantation for adult patients with Philadelphia chromosome positive acute lymphoblastic leukemia. Haematologica. 2015;100(3):392-9. PubMed | Google Scholar
- Fielding AK, Rowe JM, Buck G, Foroni L, Gerrard G, Litzow MR et al. UKALLXII/ECOG2993: addition of imatinib to a standard treatment regimen enhances long-term outcomes in Philadelphia positive acute lymphoblastic leukemia. Blood. 2014;123(6):843-50. PubMed | Google Scholar
- Coccia PF, Pappo AS, Beaupin L, Borges VF, Borinstein SC, Chugh R et al.. Adolescent and young adult oncology, version 2.2018, NCCN clinical practice guidelines in oncology. Journal of the National Comprehensive Cancer Network. 2018;16(1):66-97. Google Scholar
- Chopra A, Soni S, Verma D, Kumar D, Dwivedi R, Vishwanathan A et al. Prevalence of common fusion transcripts in acute lymphoblastic leukemia: a report of 304 cases. Asia?Pacific Journal of Clinical Oncology. 2015;11(4):293-8. PubMed | Google Scholar
- Mrózek K, Harper DP, Aplan PD. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Hematology/Oncology Clinics. 2009;23(5):991-1010. PubMed | Google Scholar
- Ajuba I, Madu A, Okocha C, Ibegbulam O, Okpala I, Nna O. Frequency and clinical impact of ETV6/RUNX1, AF4-MLL, and BCR/ABL fusion genes on features of acute lymphoblastic leukemia at presentation. Nigerian journal of clinical practice. 2016;19(2):237-41. PubMed | Google Scholar
- El-Sissy A, El-Mashari M, Bassuni W, EL-SWAAYED A. A molecular detection of BCR/ABL fusion gene in Saudi acute lymphoblastic leukemia patients. J Egypt Natl Canc Inst. 2006 Jun;18(2):109-16. PubMed | Google Scholar
- Ariffin H, Chen S-P, Kwok CS, Quah T-C, Lin H-P, Yeoh AE et al. Ethnic differences in the frequency of subtypes of childhood acute lymphoblastic leukemia: results of the Malaysia-Singapore Leukemia Study Group. Journal of Pediatric Hematology/oncology. 2007;29(1):27-31. Google Scholar
- UN. World Population Prospects: The 2017 Revision, DVD Edition. 2017. Accessed July 4, 2019.
- World Bank. Life expectancy at birth, total (years). 2017. Accessed July 4, 2019.
- Elbossaty WFM. Immunophenotypic Profile in adult patients with acute leukaemia association with clinical feature: fluorescence cytometry quantitative analysis. Journal of Molecular Biomarkers & Diagnosis. 2017;8:328.
- Daenen S, Haaxma-Reiche H, Vellenga E, Smit J, Halie R. Improved outcome of adult acute lymphoblastic leukaemia by moderately intensified chemotherapy which includes a'pre-induction'course for rapid tumour reduction: preliminary results on 66 patients. British journal of haematology. 1998;100(2):273-82. PubMed | Google Scholar
- Toft N, Schmiegelow K, Klausen TW, Birgens H. Adult acute lymphoblastic leukaemia in Denmark. A national population based retrospective study on acute lymphoblastic leukaemia in Denmark 1998-2008. British journal of haematology. 2012;157(1):97-104. Google Scholar
- Chiaretti S, Vitale A, Cazzaniga G, Orlando SM, Silvestri D, Fazi P et al. Clinico-biologic features of 5202 acute lymphoblastic leukemia patients enrolled in the Italian AIEOP and GIMEMA Protocols and stratified in age-cohorts. Haematologica. 2013:haematol. 2012.080432. Google Scholar
- WHO. The global prevalence of anaemia in 2011. Geneva: World Health Organization. 2015.